While it has been awfully quiet here for a while (has it really been three years since my last post?), I never stopped visualising data. It's just that life, uh, finds a way and time for blogging has been scarce.
I'm currently working at BioLizard 🦎 and during our most recent Christmas party, we hosted a generative art competition. I thought that my submission for this competition would be a good addition to my blog, even if it's slightly out of the ordinary data visualisation-wise. And who knows, maybe it won't take another three years now for the next update.
At BioLizard, we work a lot with sequencing data. Very basically put: you take a biological (think blood or a tumor biopsy) or environmental (think soil or sea water) sample, extract all the DNA or RNA molecules, and determine their sequence. As you may or may not know, DNA (our genetic information) is in essence built up from four different molecules or nucleotides: adenine (A), guanine (G), thymine (T), and cytosine (C). These four building blocks are strung together, one after the other, into really, really long strings, which we call chromosomes. Our longest chromosome for example, chromosome 1, is 285 MILLION nucleotides long. Small parts of these really long DNA sequences, which we call genes (of which we have, depending on the definition, between 20 and 60 thousand), are transcribed into RNA molecules. One of the main differences between DNA and RNA is that RNA contains uracil (U), instead of T. By determining the sequence of all each RNA molecule in a sample we can determine which of our genes were transcribed (or expressed) and are therefore likely to be active in our sample.
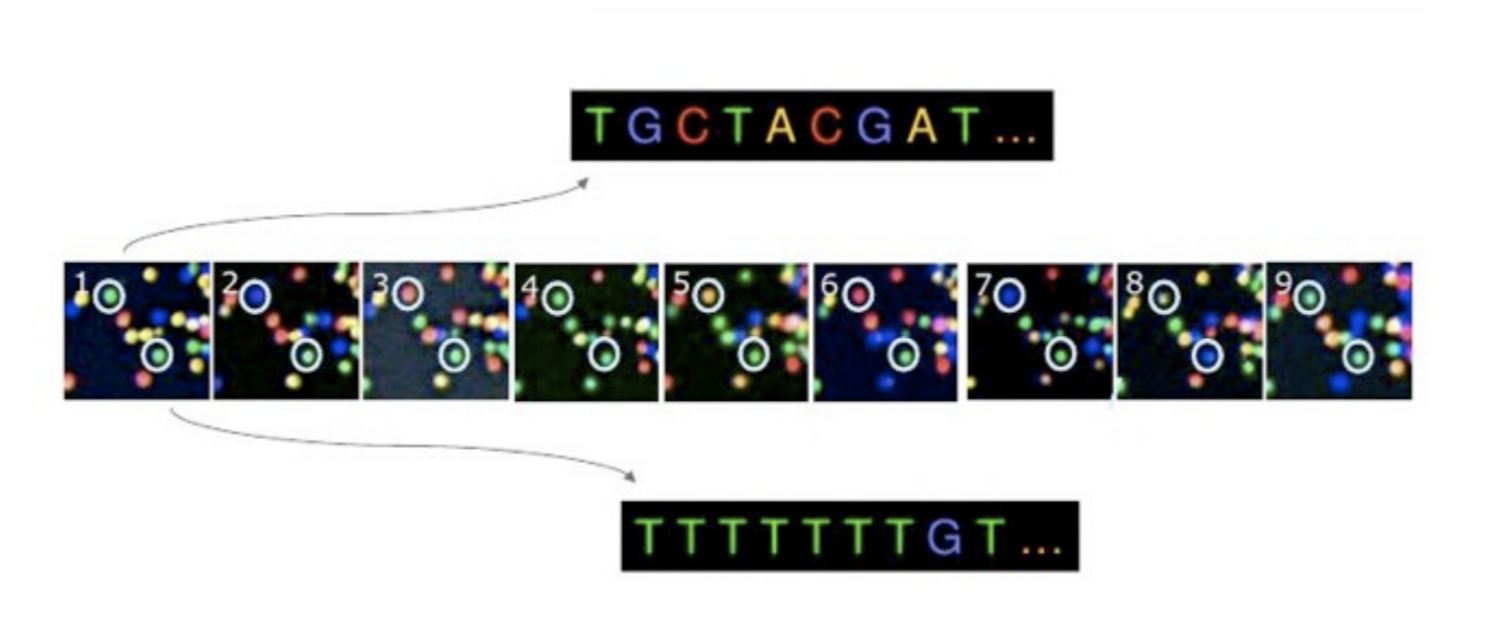
Many different technologies have been developed to do this "sequencing"[1]. Because a sample may contains hundreds of thousands of DNA or RNA molecules, a lot of effort has gone into increasing the throughput of these technologies. Most commonly used sequencing technologies rely on the use of a flow cell (title drop!). This is a small glass plate that has been densely coated with short nucleotide sequences that bind the DNA or RNA fragments that were extracted from a sample. In an iterative process, labeled nucleotides (A, T, C, and G molecules tagged with a fluorescent label) are added to the slide and washed off, building up complementary sequences to the ones that are attached to the plate. After each incorporation, a flash of light is used to excite the fluorescent labels and a sensitive camera takes a picture. Because each nucleotide label emits light with a different color, we can then string together all the pictures from the camera to determine the sequence of each DNA or RNA fragment on the slide, like this:
My idea for the generative art competition was to build a simple web app to make a sort of "flow cell selfie". The basic workflow goes like this: capture the video feed from the laptop's webcam, divide the image in a series of dots (like a mosaic with circles), get the RGB color value of the pixel at the center of each dot, convert the color to red, green, blue, or yellow (like in an actual sequencing flow cell as shown above), and plot the circles. I used the p5.js library and the final result looks something like this:
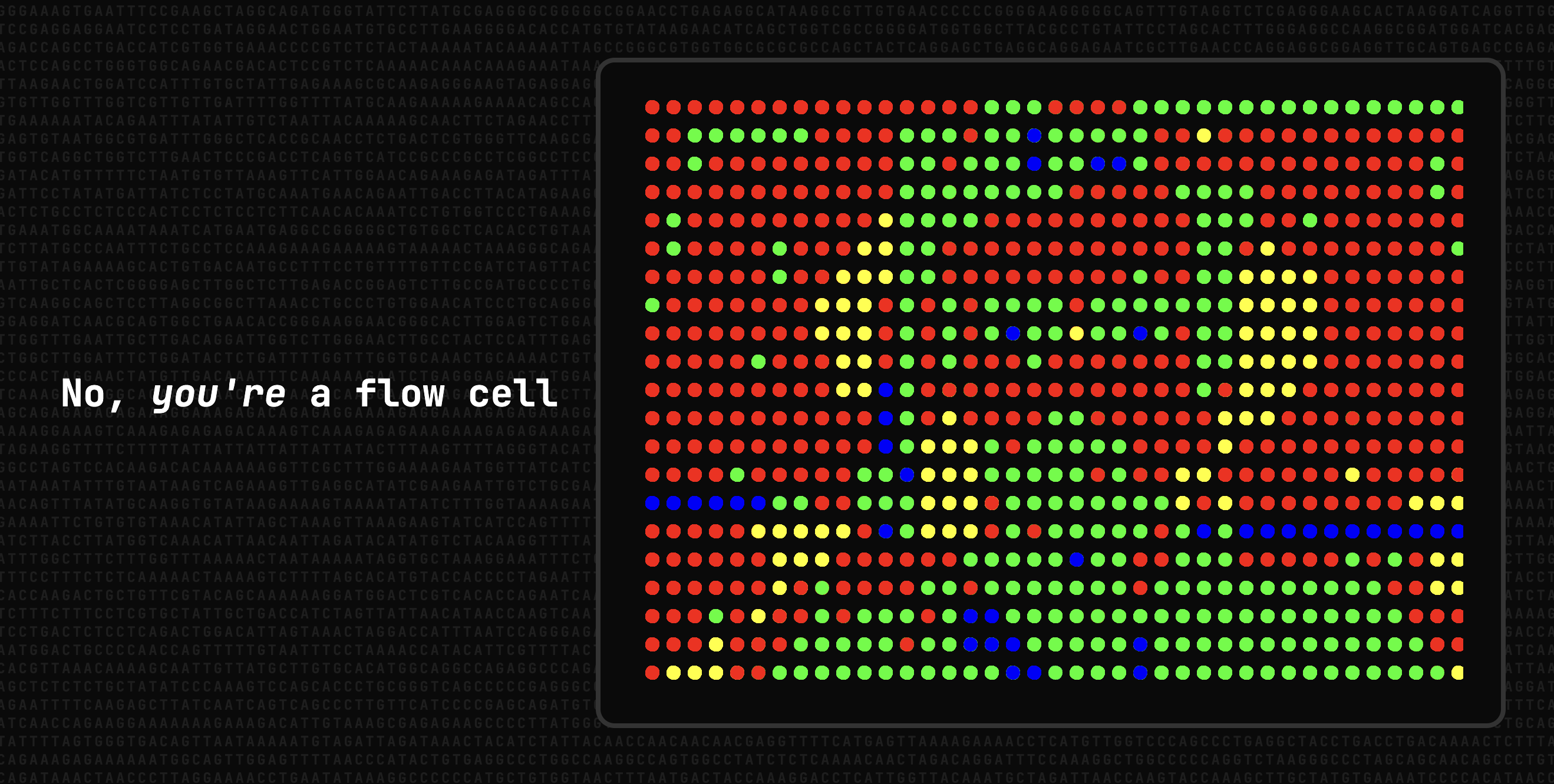
You can try the live version here. Alternatively, you can download the HTML and p5.min.js files from my GitHub repo and open the HTML file in a browser. Be aware that I have essentially done zero optimization! Increasing the resolution (more dots) crashed my browser. It's quite sluggish too, but I did not have the time to figure out how I could improve this. Happy to hear any suggestions though!
[1] My explanation of sequencing here is very brief and skips many (important) details and nuances. There are a ton of resources available online in case you want to learn more. The image I used here comes from this informative introduction to RNA-seq post from the Harvard Chan Bioinformatics Core. Illumina, a major player in the sequencing field, also has a good document that explains the basics of sequencing in general as well as their own sequencing technology. If you prefer video over text, then this step-by-step guide to DNA sequencing might be good for you. ↑ back up